Keywords: Virus structure, binding, microcalorimetry, microcalorimeter, Nano DSC, Isothermal microcalorimetry, isothermal calorimetry, Nano ITC
MC163
Introduction
COVID-19 has disrupted daily global life, and major resources are focused on disease containment, treating the sick, supporting the economy, and developing treatments and vaccines. Researchers are working to understanding the basic chemical details of viral pathogenesis, viral protein structures, and identifying therapeutics to disrupt/prevent the disease. Nano Differential Scanning Calorimetry (Nano DSC) and Isothermal Titration Calorimetry (ITC) are powerful label-free and immobilization free analytic tools for thermodynamic measurements of molecular binding, biomolecular thermostability, and enzyme kinetic reactions.
The self-assembly of viral proteins and nucleic acids to form stable particles, and the mechanisms by which viruses bind to and penetrate host cells, represent two key aspects to understanding the basis of viral infection. The thermal stability of virus coat proteins in solution, compared with the stability of the corresponding highly structured viral protein shell, can be most effectively characterized by Nano DSC, whereas fundamental information about the molecular interactions driving virus/cell binding processes can be obtained by measuring the heat evolved or absorbed during binding using ITC. This note reviews applications of Nano DSC and ITC for studying the stability of virus particles and their binding to cells, as well as for the identification of viruses.
Virus Particle Stability and Assembly
Norwalk virus is the major cause of gastroenteritis. The virus consists of an RNA genome encapsulated by a coat comprised primarily of major capsid protein, and gains entry to the body through ingestion of contaminated food or water. The major capsid protein has two structurally distinct domains and self-assembles in vitro into empty, non-infectious virus-like particles that are structurally and immunologically similar to the intact virus. The virus experiences large variations in pH as it progresses through the infective cycle. DSC studies conducted on non-infective virus-like particles (Ausar et al., 2006) between pH 3 and 8 showed two well-separated endothermic peaks at pH 3: a larger peak centered at 66 °C and a smaller peak centered at 92 °C. At less acidic pH values, the higher temperature transition shifted to lower temperatures, and at neutral pH it was superimposed on the lower temperature transition. Proteolytic studies showed that the low temperature transition was due to the proteolytically-stable large C-terminal domain, while the high temperature transition arose from the digestible smaller N-terminal domain. These results are consistent with the need for the virus to remain intact in acidic conditions following ingestion, and to disassemble and release its RNA after penetrating the gastric cell membrane and entering the (neutral pH) cytoplasm. DSC studies on capsid-protein-only shells of a parvovirus also showed different conformations and stability in response to environmental changes similar to those encountered by the virus during its life cycle (Carreira et al., 2004). In this case, conformational changes proceeded in two steps. DSC showed that heating the shells to 46 °C caused partial externalization of the N-terminus of the protein, followed by dissociation of the shell at 75 °C into monomeric proteins.
The stabilizing effect of viral nucleic acid on virus particles has been studied using intact viruses and empty coat-protein-only shells of turnip and tobacco mosaic virus. DSC and other biophysical studies of intact turnip virus showed that heat disrupts the virus in three stages: first, the RNA is released and a conformational change occurs in the shell, then the empty shell dissociates and finally, the coat protein denatures (Mutombo et al., 1993). In tobacco mosaic virus, the intact virus is stabilized by about 15 °C compared to the coat-protein-only shell, suggesting that the RNA forms specific stabilizing interactions with the protein shell (Orlov et al., 1998). The thermal stability of the coat protein alone in solution or in small aggregates is about 30 °C lower than when packed into the highly structured coat-protein-only shell. The authors point out that the presence of folded coat protein in solution is known to inhibit virus disassembly. This would aid infection, as the instability of monomeric or small coat protein aggregates would help drive the disassembly process to completion once initiated by the release and unfolding of a few coat protein molecules in the infected cell.
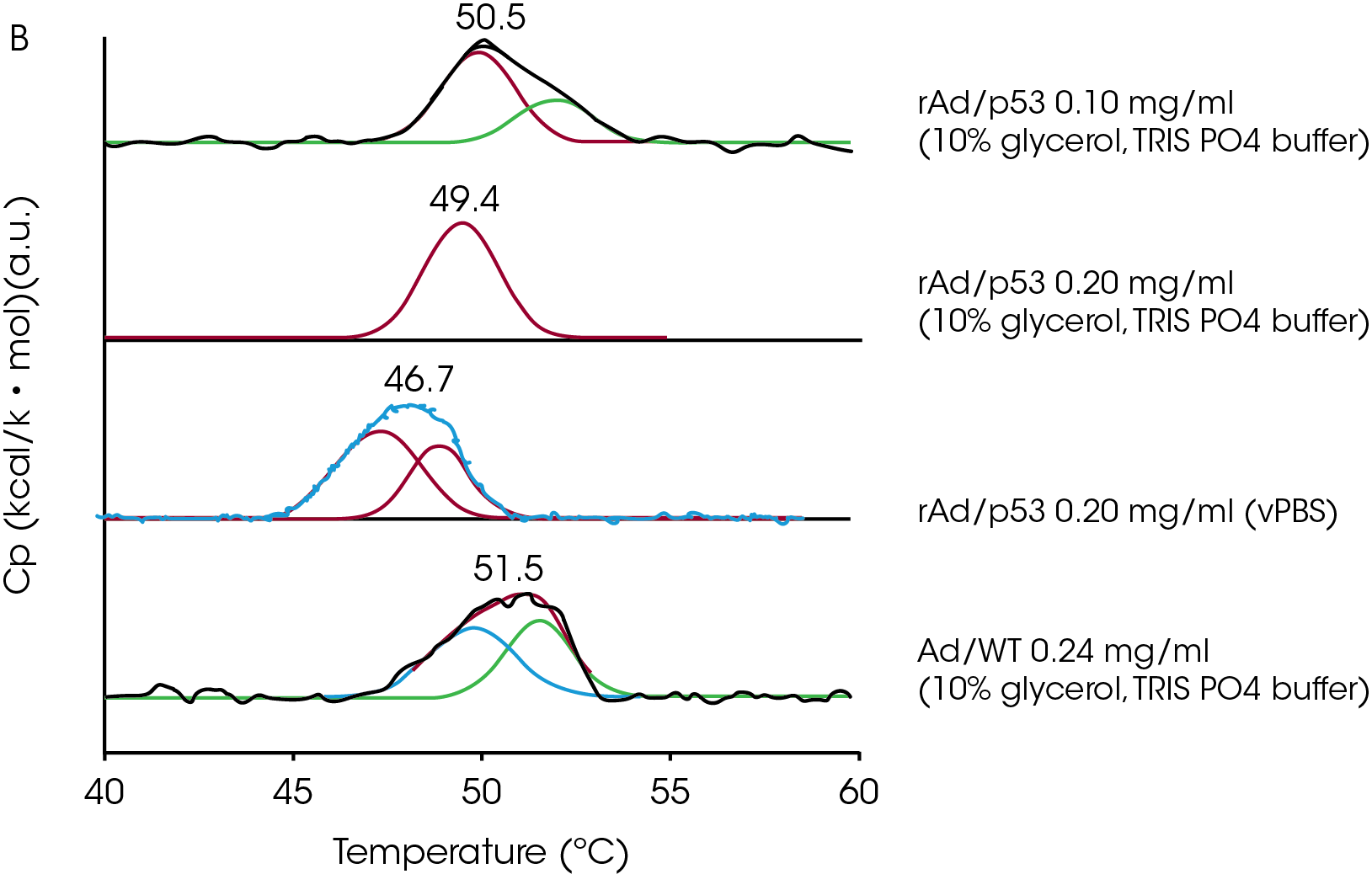
DSC has also been used to investigate the thermal stability of adenovirus, a favored vehicle for delivering transgenic material to somatic cells, with the intent of identifying the effects of environmental factors such as pH and sugars on virus stability (Rexroad et al., 2002; Ihnat et al., 2005). Ihnat et al. (2005) studied the purified wildtype major capsid protein (hexon protein), the wildtype virus (Ad/WT) and a mutant form of the virus (rAd/p53) in various buffers. The mutant virus exhibited three endothermic transitions, while the wildtype provided four transitions. The coat protein provided two endotherms (Fig A) but lacked the 50 °C endotherm characteristic of the intact virus. Deconvolution of the approx. 50 °C peak (Fig B) from the virus particles showed that this endotherm was composed of two thermal events in a concentration-dependent manner. Taken together with other calorimetric data, the authors conclude that the 50 °C transition represents the disruption of the virus particle into protein and nucleic acid, and that the transitions at approx. 70 and 75 °C arise from conformational changes in the hexon protein. The 50 °C transition is not reversible and since its position, size and shape are dependent on concentration and environment, the authors suggest that the position of this transition can be used to distinguish between adenovirus mutants, and to screen and optimize stabilizing formulations.
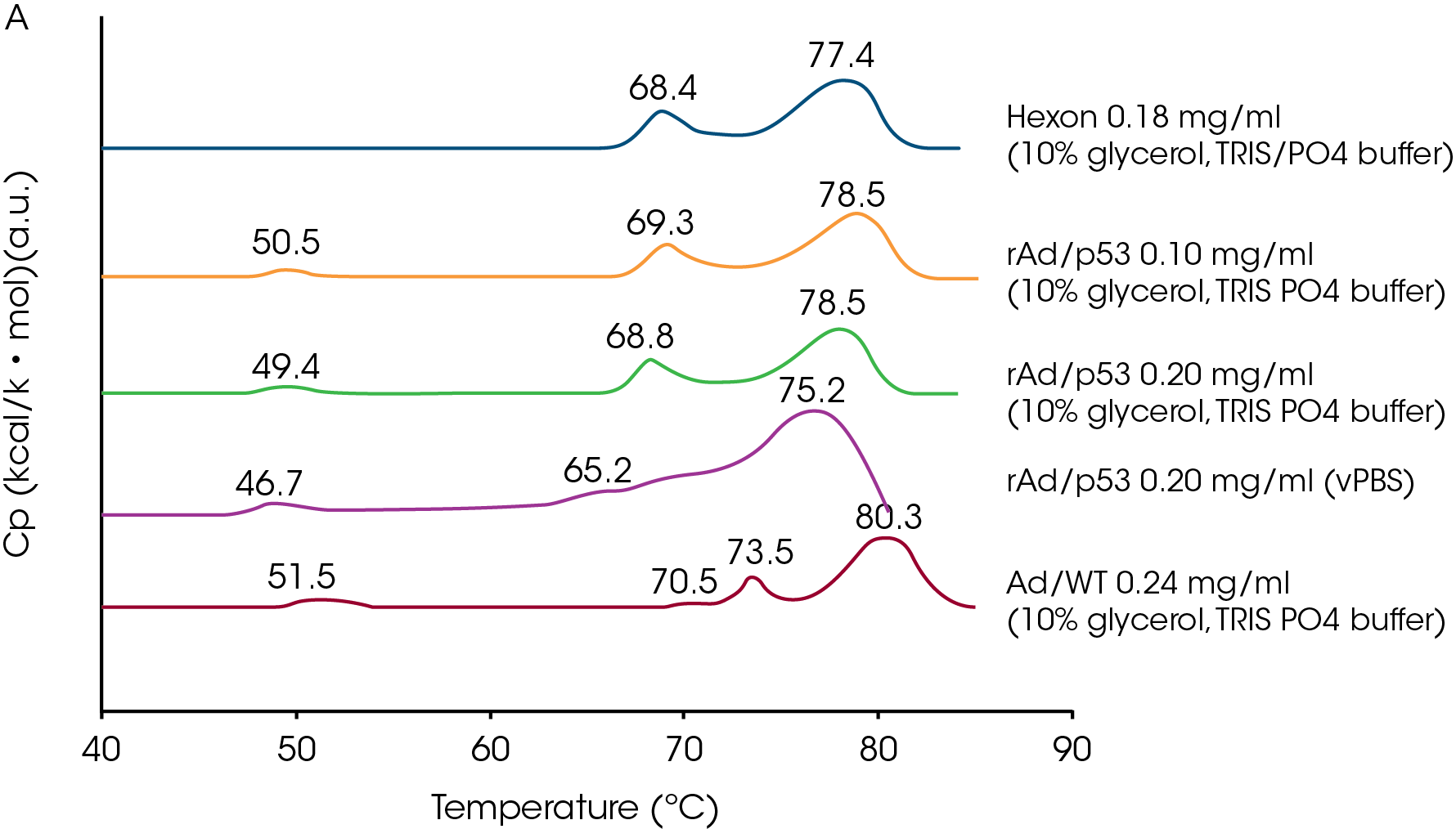
The assembly of complex viruses such as bacteriophages and some eukaryotic viruses often requires the assistance of chaperones and proteases. The thermal stability of viruses has long been studied by DSC, and changes in thermal stability during the maturation process have been correlated to structural changes observed by X-ray scattering and electron microscopy. Perhaps the most detailed thermal profiles have been obtained for the bacteriophages T4 (Ross et al., 1985; Steven et al., 1992), P22 (Galisteo and King, 1993; Galisteo et al., 1995), and HK97 (Ross et al., 2005; Ross et al., 2006). In the case of HK97, the precursor capsid protein, gp5, self-assembles into pentamers and hexamers which in turn self-assemble into procapsids. This is followed by N-terminal proteolysis, expansion of the capsid particle, conformational rearrangements, covalent cross-linking, and insertion of the DNA (Ross et al., 2006). The maturation process results in progressively more stable particles, and some steps (proteolysis and cross-linking) prevent reversal of the process. The intact virus cannot be thermally disassembled without the use of 5 M urea.
Virus Entry into Cells
Viruses enter cells through one or a combination of two general mechanisms: direct fusion with the cell membrane (enveloped viruses) and entry via endocytotic or other vesicles (non-enveloped viruses). The latter mechanism is mediated by cell surface receptors, although some viruses may fuse with the host cell membrane in the absence of a receptor. Calorimetry has been used to study both types of infection pathways.
Enveloped animal viruses enter the host cell by fusing with the host cell membrane, induced by conformational changes in viral coat glycoproteins. The conformational change can be triggered by the acidic environment of the endosome, or by interaction with a specific receptor on the cell surface. Influenza virus fuses with cell membranes under acidic conditions. The virus coat consists of a lipid membrane and three transmembrane proteins of which one, hemagglutinin, undergoes a large conformational change at acidic pH. This exposes a hydrophobic segment that drives virus fusion with the host cell membrane. DSC of purified hemagglutinin at neutral pH produces a single large-enthalpy cooperative endotherm with a Tm of 66 °C, whereas at pH 5 the protein unfolds in a less cooperative manner at 45 °C, with approximately one-third the enthalpy change (Remeta et al., 2002). DSC and spectroscopic studies on intact influenza virus, conducted to determine how the hemagglutinin protein responds to pH and temperature when embedded in the viral membrane, also showed significant differences between acidic and neutral pH (Epand and Epand, 2002). As with the purified protein, hemagglutinin in the intact virus unfolded cooperatively at about 66 °C at neutral pH, but at pH 5, the Tm decreased only to 60 °C and the enthalpy remained essentially unchanged, although cooperativity was significantly reduced. The authors concluded that the decreased stability of hemagglutinin at acidic pH initiates virus/cell membrane fusion mediated by the exposed hydrophobic segment, but that hemagglitinin within the intact virus is less destabilized than the purified protein under similar conditions. ITC experiments with intact and inactivated influenza virus at neutral and acid pH allowed binding and membrane fusion events to be separated and showed that membrane fusion is an endothermic reaction occurring exclusively at acidic pH (Nebel et al., 1995).
ITC and other studies on vesicular stomatitis virus showed that the viral fusion protein bound to and fused with phosphatidyl choline/phosphatidyl serine (1:3) vesicles exothermally at pH 6.0, whereas binding but no fusion occurred at pH 7.5 (Carneiro et al., 2002, 2006). The fact that binding was exothermic suggested that it might be driven by electrostatic interactions. No binding was observed when the vesicles did not contain phosphatidyl serine, or when the histidines in peptides corresponding to the putative binding region on the viral fusion protein were replaced with alanines. The ITC data suggest that binding of the viral fusion protein to the cell brings histidine residues in the binding region of the protein in close proximity with the negatively charged phosphatidyl serine head groups, leading to protonation of the histidines and interaction of the fusion protein with the cell membrane.
HIV is perhaps the most extensively studied enveloped virus. Infection is initiated when the viral coat glycoprotein, gp120, interacts with CD4 receptors on the target cell, triggering conformational changes in the virus envelope and leading to fusion with the target cell. ITC studies of the interaction between gp120 and CD4 (Myszka et al., 2000) showed a very favorable binding enthalpy (approx. -63 kcal/mol (-264 kJ/mol)), indicating the formation of a large number of hydrogen bonds and van der Waals interactions. However, the binding entropy is large and unfavorable, indicating significant loss of conformational freedom upon binding and resulting in medium strength (2 x 108 M-1) binding. In addition, the binding reaction is slow. Independent studies on the CD4 protein showed that its structure remains essentially unchanged upon binding, demonstrating that the unfavorable entropy arises primarily from structural rearrangement of gp120. Slow binding is consistent with significant conformational changes occurring during complex formation.
Extensive studies on a nonenveloped virus, simian virus 40, has shed light on how a virus recognizes and binds to its receptor on the host cell, without binding so tightly that replicated viruses cannot migrate to new host cells. ITC data showed that the virus recognizes its glycoprotein receptor with millimolar affinity, but glycan screening experiments showed that when presented with an array of glycans, the virus binds preferentially to the branched oligosaccharide found on the receptor. Crystallographic data show that binding specificity is achieved by the two branches of the oligosaccharide interacting simultaneously with the binding cleft (Neu et al., 2008).
Identifying Viruses by DSC
In 1997, Shnyrov et al. showed that scanning a Newcastle disease virus sample to successively higher temperatures on DSC (successive annealing) provided four endothermic two-state transitions which corresponded to the four major proteins in the virus. The results suggested that the DSC profile of a virus could be used as a means of identification. This concept has been extended beyond the identification of the species of virus, to the identification of individual strains. For example, Krell et al. (2005) used DSC to characterize various polio and influenza virus strains. Each strain had a characteristic pattern of multiple unfolding transitions which differed in Tm, shape and enthalpy, with significant pattern differences even between closely-related strains (e.g., the highly-related H1N1 influenza strains A/PR8/34 and New Caledonia, or the H3N2 strains X-31 and Panama). The authors suggest that the surface coat proteins strongly influence the DSC profile of the virus. Since it is these proteins that often differ between closely related strains, DSC could be a widely-applicable, simple and fast clinical approach for identifying infective viral strains.
Summary
The body of research incorporating DSC and ITC studies for understanding the structure and mode of action of viruses is growing rapidly. DSC is invaluable for elucidating the thermodynamic profile of a virus structure, leading to an understanding of the stability of viral components and the reliable characterization and identification of virus strains. Virus virulence depends on the ability of the virus to specifically bind to its target and infect the cells. ITC has been shown to be a robust assay technology that can be used to effectively characterize any inter-molecular binding events occurring between a virus and its receptors. Because viruses are composed of only a few major protein and nucleic acid components, it is perhaps not surprising that details of how these components pack and interact with each other can be studied calorimetrically using intact viruses. At the same time, the level of specific structural information from a well-designed DSC or ITC experiment conducted on intact viruses can rival that of other techniques that require purified protein or nucleic acid components to provide interpretable data. Rapid accurate evaluation of viral structure and virulence factors could be critical for expanding the structure-activity relationship database for viruses, thereby allowing for a timely response to viral outbreaks and pandemics.
References
- Ausar, S., T. Foubert, M. Hudson, T. Vedvick and C. R. Middaugh (2006). Conformational stability and disassembly of Norwalk virus-like particles. J. Mol. Biol. 281, 19478-19488.
- Carneiro, F, M. L. Bianconi, G. Weissmuller, F. Stauffer and A. Da Poian (2002). Membrane recognition by vesicular stomatitis virus involves enthalpy-driven protein-lipid interactions. J. Virol. 76, 3756-3764.
- Carneiro, F. et al. (2006). Probing the interaction between vesicular stomatitis virus and phosphatidylserine. Eur. Biophys. J. 35, 145-154.
- Epand, R. M. and R. F. Epand (2002). Thermal denaturation of influenza virus and its relationship to membrane fusion. Biochem. J. 365, 841-848.
- Carreira, A., M. Menendsz, J. Reguera, J. M. Almendral and M. Mateu (2004). In vitro disassemably of a parvovirus capsid and effect on capsid stability of heterologous peptide insertions in surface loops. J. Mol. Biol. 279, 6517-6525.
- Galisteo, M. L. and J. King (1993). Conformational transformations in the protein lattice of bacteriophage P22 procapsids. Biophys. J. 65, 227-235.
- Galisteo, M. L., C. L Gordon and J. King (1995) Stability of wild-type and temperature-sensitive protein subunits of the phage P22 capsid. J. Biol. Chem. 270, 16595-16601.
- Ihnat, P. et al. (2006). Comparative thermal stabilities of recombinant adenoviruses and hexon protein. Biochim. Biophys. Acta 1726, 138-151.
- Krell, T., et al., (2005) Characterization of different strains of poliovirus and influenza virus by differential scanning calorimetry. Biotechnol. Appl. Biochem. 41, 241-256.
- Mutombo, K., B. Michels, H. Ott, R. Cerf and J. Witz (1993). The thermal stability and decapsidation of tymoviruses: a differential scanning calorimetric study. Biochimie 75, 667-674.
- Myszka, D. G. et al. (2000). Energetics of the HIV gp120-CD4 binding reaction. PNAS 97, 9026-9031.
- Nebel, S., I. Bartoldus and T. Stegmann (1995). Calorimetric detection of influenza virus induced membrane fusion. Biochemistry 34, 5705-5711.
- Neu, U., K. Woeliner, G. Gauglitz and T. Stehle (2008). Structural basis of GM1 ganglioside recognition by simian virus 40. PNAS 105, 5219-5224.
- Orlov, V. N., S. V. Kust, P. V. Kalmykov, V. P. Krivosheev, E. N. Dobrov and V. A. Drachev (1998). A comparative differential scanning calorimetric study of tobacco mosaic virus and of its coat protein ts mutant. FEBS Letters 433, 307-311.
- Remeta, D. P., M. Krumbiegel, C. Minetti, A. Puri, A. Ginsburg and R. Blumenthal (2002). Acid-induced changes in thermal stability and fusion activity of influenza hemagglutinin. Biochemistry 41, 2044-2054.
- Rexroad, J., C. Wiethoff, A. Green, T. Kierstead, M. Scott and C. R. Middaugh (2002). Structural stability of adenovirus type 5. J. Pharma. Sci., 92, 665-678.
- Ross, P. D., L. W. Black, M. E. Bisher and A. C. Steven (1985). Assembly-dependent conformational changes in a viral capsid protein. Calorimetric comparison of successive conformational states of the gp23 surface lattice of bacteriophage T4. J. Mol. Biol. 183, 353-364.
- Ross, P., N. Cheng, J. Conway, B. Firek, R. Hendrix, R. Duda and A. Steven (2005). Crosslinking renders bacteriophage HK97 capsid maturation irreversible and effects an essential stabilization. EMBO J., 24, 1352-1363.
- Ross, P., J. Conway, N. Cheng, L. Dierkes, B. Firek, R. Hendrix, A. Steven and R. Duda (2006). A free energy cascade with locks drives assembly and maturation of bacteriophage HK97 capsid. J. Mol. Biol. 364, 512-525.
- Steven, A. C., H. L. Greenstone, F. P. Booy, L. Black and P.
D. Ross (1992). Conformational changes of a viral capsid protein. Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, co-operativity, and super-stabilization by Soc binding. J. Mol. Biol. 228, 870-884. - Shnyrov, V. L., G. G. Zhadan, C. Cobaleda, A. Sagrera, I. Munoz-barroso and E. Villar (1997). A differential scanning calorimetric study of Newcastle disease virus: identification of proteins involved in thermal transitions. Archives Biochem. Biophysics 341, 89-97.
Acknowledgement
Click here to download the printable version of this application note.

