Keywords: Proteins, amino acids, DSC
MC175
Fifty years ago, the elegant experiments of Christian Anfinsen showed unequivocally that the linear sequence of amino acids of a protein (its primary structure) encodes all the information for its three-dimensional structure (Sela et al., 1957; Anfinsen, 1973). It is now clear that the sequence directs the interactions both between component amino acids, and between the protein and the solvent, and that these interactions are governed by thermodynamics. What is not clear is how the sequence encodes the complex structure of a protein.
The thermodynamic balance in proteins is delicate: the folded structure of a small protein is stabilized by approximately 80‑120 kJ/mol (20-30 kcal/ mol) compared to its unfolded structure, and many proteins thermally unfold by 70 °C. If most proteins can be denatured by relatively small increases in temperature, and if the balance between folded and unfolded states is held by the equivalent of 5 – 8 hydrogen bonds, how are thermophilic proteins stabilized, allowing organisms to thrive at elevated pressures and at temperatures above 100 °C (Perl and Schmid, 2002)? In the absence of thermal changes, what thermodynamic events trigger proteins that cause amyloid and prion diseases to switch from one stable conformation to another, with fatal consequences (Mastrianni, 2004)? Would a thorough comprehension of the interactions controlling conformation permit the full complement of three-dimensional protein structures to be accurately predicted from an organism’s genome (Webster 2000)? Achieving a fundamental understanding of the thermodynamics directing the formation of macromolecular noncovalent bonds would answer these questions, and critically impact such diverse endeavors as the self-assembly of protein inspired supramolecular structures (Yeates and Padilla, 2002), the composition of shape-memory biocompatible polymers (Langer and Tirrell, 2004), the rational design of protein-specific small molecule drugs (Waldron and Murphy, 2003) and the formulation of stable protein therapeutics (Remmele and Bombotz, 2000).
The thermodynamic parameters that control noncovalent bond formation in proteins and other macromolecules can be directly studied and quantified by differential scanning calorimetry (DSC). This Application Note examines the utility of DSC for characterizing protein stability.
Analyzing Protein Thermal Stability
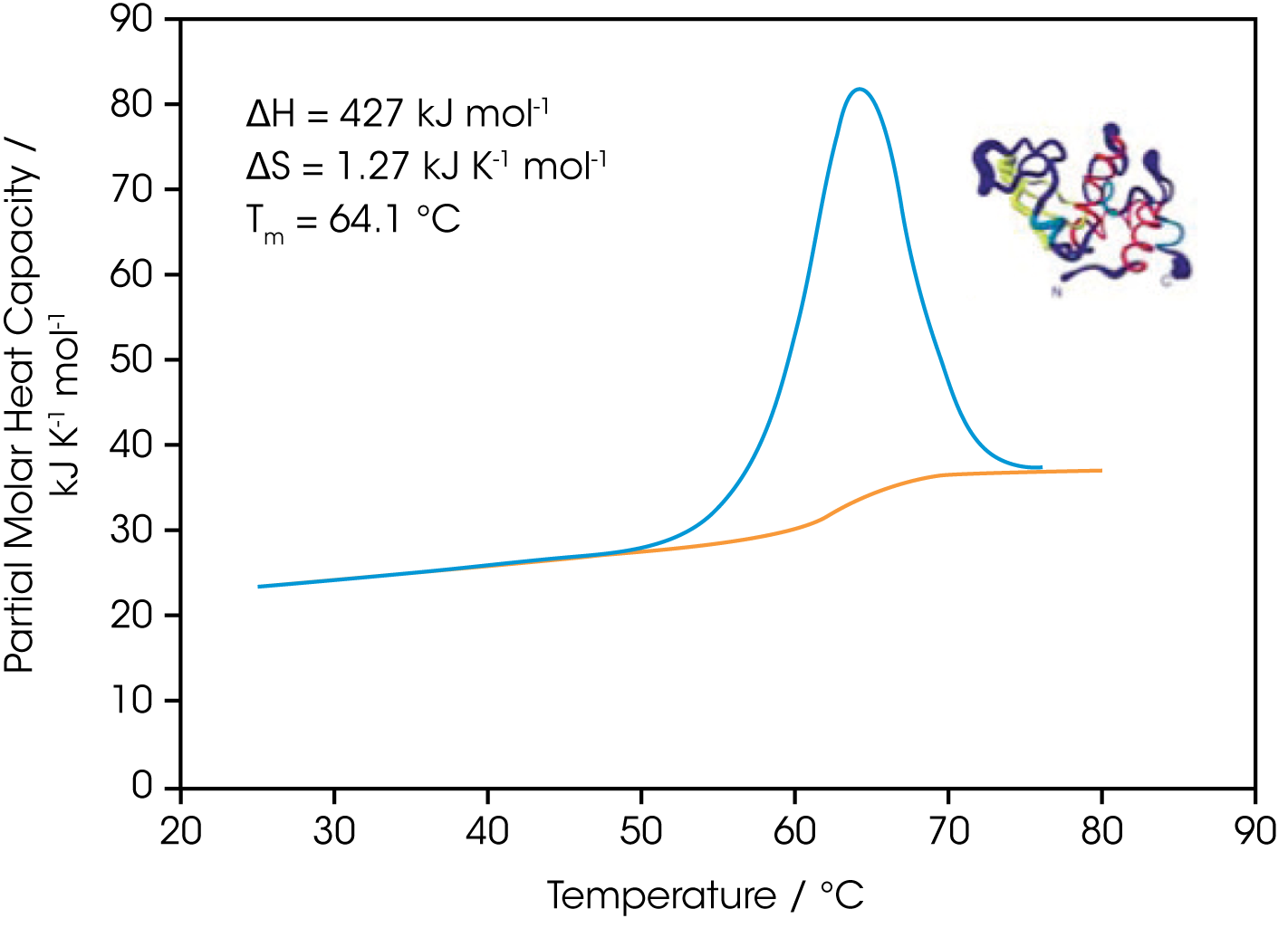
As described in the DSC Overview Note, analyzing the stability of a protein in dilute solution involves determining changes in the partial molar heat capacity of the protein at constant pressure (ΔCp). The change in heat capacity of a compound reflects its ability to absorb heat and experience a defined increase in temperature. Because of its extensive hydrogen bond network, water has a substantially higher heat capacity than organic compounds such as proteins. In addition, water is more ordered and tightly packed adjacent to hydrophobic patches on the protein’s surface: since water cannot make hydrogen bonds to nonpolar moieties, hydrogen bonding between water molecules is maximized (clathrate formation) at the protein-solvent interface (Shinoda, 1977). As the temperature is raised, the ordered water shell around the protein starts to disorder and become more like bulk water. This ‘melting’ produces the anomalously large heat capacity of protein aqueous solutions. The contribution of the protein to the calorimetrically measured heat capacity (its partial Cp) is p determined by subtracting a scan of a buffer blank from the sample data prior to analysis. The partial Cp is not just the heat capacity of the protein, but also includes any effects the protein had on the solvent, such as promoting clathrate formation (Freire, 1995). Heating the protein sample initially produces a slightly increasing baseline but as heating progresses, heat is absorbed by the protein and causes it to thermally unfold over a temperature range characteristic for that protein, giving rise to an endothermic peak (Fig. 1 ). During the unfolding process, water molecules around the protein reorganize and restructure as more non-polar sidechains are exposed. Once unfolding is complete, heat absorption decreases, and a new baseline is established.
After blank subtraction, the data can be analyzed to provide a complete thermodynamic characterization of the unfolding process. Integration of the heat capacity of the sample vs. temperature yields the enthalpy (ΔH)
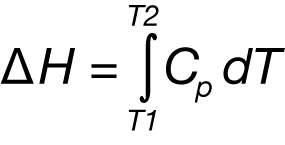
of the unfolding process, which is due to endothermic events such as the breaking of hydrogen bonds, and exothermic processes such as the disruption of hydrophobic interactions (Bruylants et al., 2005). The shift in baseline before and after the transition represents the change in heat capacity (ΔCp of the protein (and associated water) caused by unfolding. The transition midpoint (Tm, or the melting temperature) is the temperature at which half the protein molecules are folded and half are unfolded. The entropy (ΔS) is obtained from the area under the curve of Cp/T vs. T.
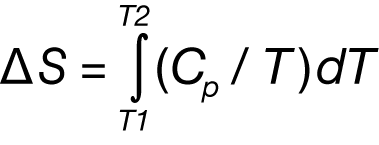
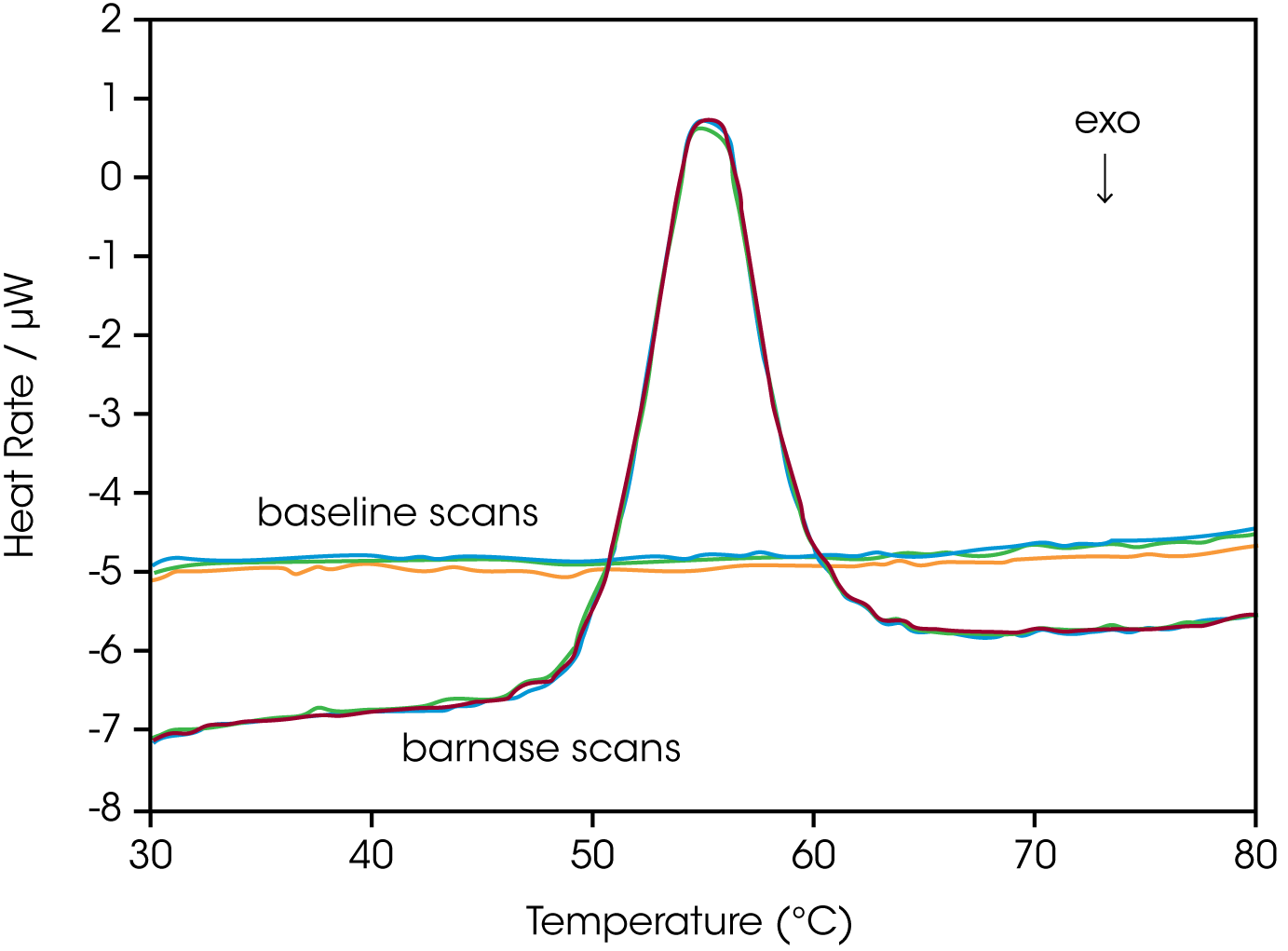
Establishing the reversibility of the unfolding reaction is not necessary if only the determination of Tm is of interest. However, a full thermodynamic characterization of the unfolding event requires that the reversibility of the unfolding transition be verified (Marky and Breslauer, 1987). This is accomplished by rescanning the sample under identical conditions and obtaining a second endotherm that is superimposable on the first. Irreversibility, if observed, may be due to a second (irreversible) unfolding event occurring after the first (reversible) unfolding transition at Tm. The capillary cells in the DSC delay the onset of irreversible protein aggregation and precipitation, allowing a complete, interpretable scan to be obtained on essentially any sample. For example, 100 μM bovine serum albumin (in 0.1 M phosphate buffer, pH 7.2, scanned from 20 to 90 °C at 1 °C/min) precipitated after a single scan, whereas under identical conditions precipitate was not observed below 300 μMin in the DSC (capillary cells).
The sharpness of the transition peak is indicative of the cooperative nature of the unfolding process. If unfolding produces a narrow, relatively symmetric peak such as in Fig. 2, the transition is likely to be two-state, reversible and highly cooperative (Bruyants et al., 2005). This is often the situation for small, single domain proteins; the more complex behavior of multidomain proteins is considered in detail in a separate application note. Two-state behavior can be verified by fitting the curve to the two-state van’t Hoff equation; this calculation is performed by software supplied with DSC. The van’t Hoff enthalpy obtained (ΔHvH) can be different from the calorimetric enthalpy since the latter includes all contributions from the sample, including solvent rearrangement. If the van’t Hoff and calorimetric enthalpies are the same, the denaturation process may be accurately approximated by the two-state model. If the van’t Hoff enthalpy is smaller than the calorimetric enthalpy, an unfolding intermediate is likely formed, while if it is larger than the calorimetric enthalpy, the protein is likely multimeric (Marky and Breslauer, 1987).
Correlating Thermodynamic Properties to Stability
The stability of a protein (and hence its Tm) depends on environmental conditions. For example, a protein will be optimally stable at a characteristic pH and will be destabilized at pH values deviating from that optimum (Privalov and Khechinashvili, 1974). The conformational stability of a protein may be affected by its concentration (showing the importance of protein-protein interactions), ionic strength of the solution (showing the role of electrostatic interactions) and scan rate (showing that thermal denaturation has a kinetic component). A complete thermodynamic characterization of a simple protein such as lysozyme illustrates the complex interplay of stabilizing and destabilizing interactions and shows that lysozyme is unusually stable for a small protein (Pfeil and Privalov, 1976; Griko et al., 1995). The different heat capacities of the folded and denatured form of the protein show that the enthalpies and entropies of both states are strongly temperature dependent, but they largely compensate each other so that the net stability of a typical small protein is approximately 80-120 kJ/mol (20-30 kcal/mol). The large ΔCp of unfolding also indicates p that a protein will have maximum stability at a given temperature (depending on its sequence, pH, etc.), and that its net stability will decrease above and below that temperature. Thus, proteins can not only be denatured by heating but can also be denatured by cooling.
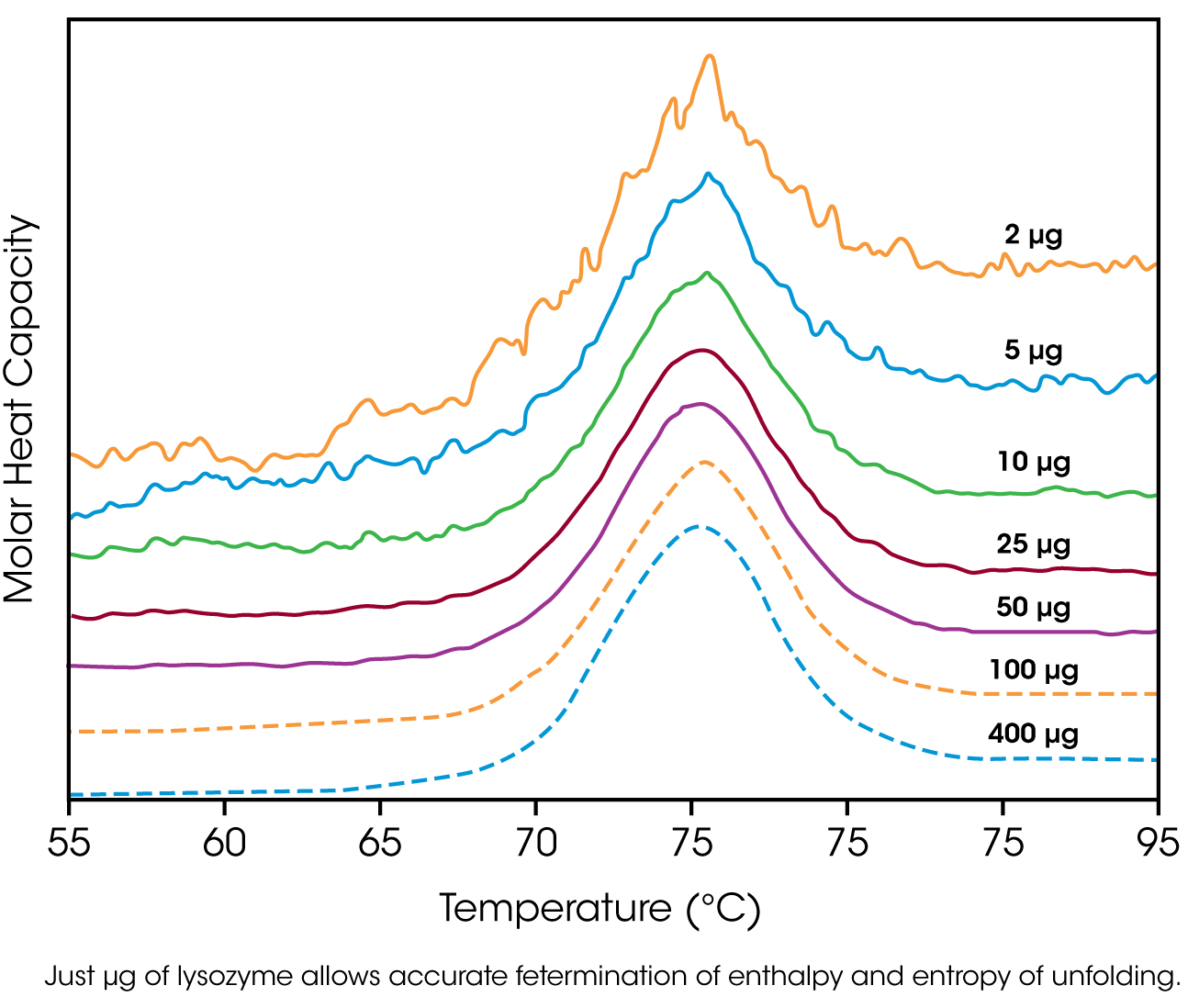
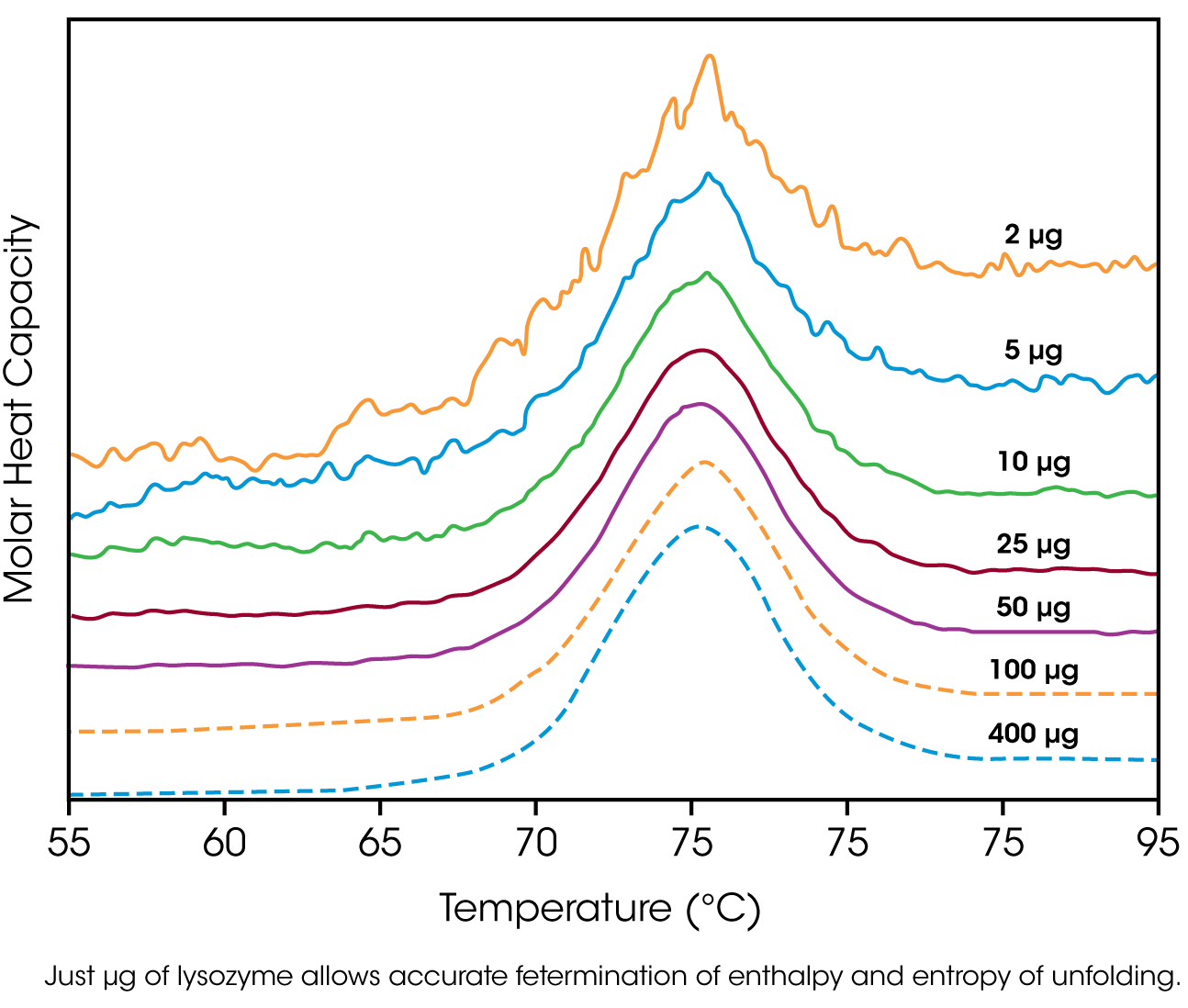
The amount of protein available for characterization can be very limited, which in the past restricted the application of biophysical techniques such as DSC. Recent advances in instrument sensitivity have alleviated this constraint by significantly decreasing the amount of sample required for DSC analyses. Fig. 3 shows raw data for lysozyme scanned on a DSC. Protein amounts ranged from 400 μg to 2 μg in the 300 μL sample cell. Thermodynamic parameters obtained at all concentrations are consistent (Table 1), showing that even 2 μg of protein can be sufficient to allow accurate thermodynamic characterization of a protein.
Can the thermodynamic properties of a protein, analyzed by DSC, be correlated to structure and stability? As more proteins are carefully characterized both experimentally and computationally, correlations between thermodynamics and structure are being revealed, albeit not yet at a reliably predictive level. A discussion of the stabilizing and destabilizing interactions outlined above comprises a vast body of literature; much of this literature has been rigorously and comprehensively reviewed by Robertson and Murphy (1997).
Table 1. Thermodyanamic parameters obtained from the lysozyme scans presented in Figure 3.
| Lysozyme in cell (μg) | Calorimetric | van’t Hoff | ||
|---|---|---|---|---|
| ΔH (kJ mol-1) | ΔS (kJ K-1 mol-1) | Tm (°C) | ΔH (kJ mol-1) | |
| 400 | 512 | 1.46 | 78.0 | 515 |
| 100 | 512 | 1.46 | 78.0 | 509 |
| 50 | 517 | 1.47 | 77.9 | 513 |
| 25 | 513 | 1.46 | 77.8 | 513 |
| 10 | 515 | 1.47 | 78.0 | 515 |
| 5 | 490 | 1.40 | 78.1 | 510 |
| 2 | 503 | 1.43 | 77.8 | 499 |
Rationalizing the Stabilities of Folded Conformations: Mutant Vs. Wild-type Proteins
Hydrophobicity scales of the amino acids, and propensity scales for amino acids in various secondary structures (Koehl and Levitt, 1999; Acevedo and Lareo, 2005), are used in conjunction with molecular modeling to predict the effect of specific mutations. Systematic mutation of proteins with known three-dimensional structures often aims to quantify the resultant stability changes in terms of the energetics of unfolding. A single site-specific mutation can give rise to a large increase or decrease in stability, but many mutations produce only small effects. The ability of proteins to make subtle solvation and conformational changes in order to compensate for the enthalpic and entropic disturbances caused by a mutation can result in only small free energy changes being observed upon thermal denaturation of the protein. Nonetheless, the growing database of proteins thoroughly characterized by DSC is aiding comprehension of mutational effects on the stability of proteins, although care must be exercised in the analyses (Sturtevant, 1994; Plum and Breslauer, 1995; Robertson and Murphy, 1997; Brylants et al., 2005). The recent redesign of proteins for increased stability (Fowler et al., 2005; Lee et al., 2005), and in particular the rational redesign of enzymes for both enhanced entropic stability and catalytic function (O’Fagain, 2003; Eijsink et al., 2004), illustrate that as long as the mechanisms controlling the stability and thermal unfolding of a protein are sufficiently understood, it is possible to logically and predictably alter the properties of proteins of known structure.
Summary
Understanding the thermodynamics controlling the formation of macromolecular noncovalent bonds is necessary if the promise of diverse endeavors such as the design of supramolecular assemblies and therapeutic proteins are to be realized. Many factors are responsible for the stability of a protein, including hydrogen bonds to the solvent and intramolecular interactions. Rigorous characterization of the thermodynamic basis of a protein’s stability is prerequisite for enhancing its thermal and functional characteristics by sequence alteration. DSC is the most direct approach for determining the intrinsic stability of a protein in dilute solution, allowing the relationship between enthalpy and entropy of the denaturation process to be quickly established. The superior sensitivity of the CSC N-DSC II or N-DSC Ill allows accurate thermodynamic parameters to be obtained from just a few micrograms of protein.
References
(Preference has been given to current references. Citation does not imply that a paper is necessarily the original reference to a study.)
- Acevedo, O. E. and L. R. Lareo. (2005) Amino acid propensities revisited. OMICS 9, 391-399.
- Anfinsen, C. B. (1973) Principles that govern the folding of protein chains. Science 181, 223-230.
- Bruylants, G., J. Wouters and C. Michaux. (2005) Differential scanning calorimetry in life sciences: thermodynamics, stability, molecular recognition and applications in drug design. Curr. Med. Chem. 12, 2011-2020.
- Chan, H. S., S. Shimizu and H. Kaya. (2004) Cooperativity principles in protein folding. Methods Enzymol. 380, 350- 379.
- Cooper, A., M. A. Nutley and A. Wadood. (2001) Differential scanning calorimetry. p. 287-318. In S. E. Harding and B. Z. Chowdry (Eds.) Protein-Ligand Interactions: hydrodynamics and calorimetry. Oxford University Press, Oxford.
- Eijsink, V. G. H. et al. (2004) Rational engineering of enzyme stability. J. Biotechnol. 113, 115-120.
- Fersht, A. R. and L. Serrano. (1993) Principles of protein stability derived from protein engineering experiments. Curr. Opin. Struct. Biol. 3, 75-83.
- Fowler, S. B. et al. (2005) Rational design of aggregationresistant bioactive peptides: reengineering human calcitonin. PNAS 102, 10105-10110.
- Freire, E. (1995) Thermal denaturation methods in the study of protein folding. Methods Enzymol. 259, 144-168.
- Griko, Y. V., E. Freire, G. Privalov, H. Van Dael and P. L. Privalov. (1995) The unfolding thermodynamics of c-type lysozymes: a calorimetric study of the heat denaturation of equine lysozyme. J. Mol. Biol. 252, 447-459.
- Koehl, P. and M. Levitt. (1999) Structure-based conformational preferences of amino acids. PNAS 96, 12524-12529.
- Langer, R. and D. A. Tirrell. (2004) Designing materials for biology and medicine. Nature 428, 487-492.
- Lee, C-F., G. I. Makhatadze and K-B Wang. (2005) Effects of charge-to-alanine substitutions on the stability of ribosomal protein L30e from Thermococcus celer. Biochemistry 44, 16817-16825.
- Marky, L. A. and K. J. Breslauer. (1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting
curves. Biopolymers 26, 1601-1620. - Mastrianni, J. A. (2004) Prion diseases. Clinical Neurosci. Res. 3, 469-480.
- Mendes, J., R. Guerois and L. Serra no. (2002) Energy estimation in protein design. Curr. Opin. Struct. Biol. 12, 441-446.
- O’Fagain, C. (2003) Enzyme stabilization: recent experimental progress. Enzyme Microb. Technol. 33, 137-149.
- Perl, D. and F. X. Schmid. (2002) Some like it hot: the molecular determinants of protein stability. ChemBiochem. 3, 39-44.
- Pfeil, W. and P. L. Privalov. (1976) Thermodynamic investigations of proteins Ill. Thermodynamic description of lysozyme. Biophys. Chem. 4, 41-50.
- Plum, G. E. and K. J. Breslauer. (1995) Calorimetry of proteins and nucleic acids. Curr. Opin. Struct. Biol. 5, 682-690.
- Privalov, P. L. and N. N. Khechinashvili. (1974) A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J. Mol. Biol. 86, 665-684.
- Privalov, G .P. and P. L. Privalov. (2000) Problems and perspectives in microcalorimetry of biological macromolecules. Methods Enzymol. 323, 31-62.
- Remmele, R. L. and W. R. Gombotz. (2000) Differential scanning calorimetry: A practical tool for elucidating stability of liquid pharmaceuticaIs. BioPharm. 13, 36-46.
- Robertson, A. D. and K. P. Murphy. (1997) Protein structure and the energetics of protein stability. Chem. Rev. 97, 1251-1267.
- Russ, W. P. and R. Ranganathan. (2002) Knowledge-based potential functions in protein design. Curr. Opin. Struct. Biol. 12, 447-452.
- Scharnagl, C., M. Reif and J. Friedrich. (2005) Stability of proteins: temperature, pressure and the role of solvent. Biochim. Biophys. Acta. 1749, 187-213.
- Sela, M., F. W. White and C. B. Anfinsen. (1957) Reductive cleavage of disulfide bridges in ribonuclease. Science 125, 691-692.
- Shinoda, K. (1977) ‘Iceberg’ formation and solubility. J. Phys. Chem. 81, 8069-8072.
- Sturtevant, J. M. (1994) The thermodynamic effects of protein mutations. Curr. Opin. Struct. Biol. 4, 69-78. 30.
- Yeates, T. O. and J. E. Padilla. (2002) Designing supramolecular protein assemblies. Curr. Opin. Struct. Biol. 12, 464-470.
- Webeter, D. M. (Ed.) (2000) Protein structure prediction. Humana Press.
Click here to download the printable version of this application note.

